2020.03.05
Rearrangement analysis of multiple bacterial genome
SOKENDAI Student Dispatch Program program year: 2019
Genetics Mehwish Noureen
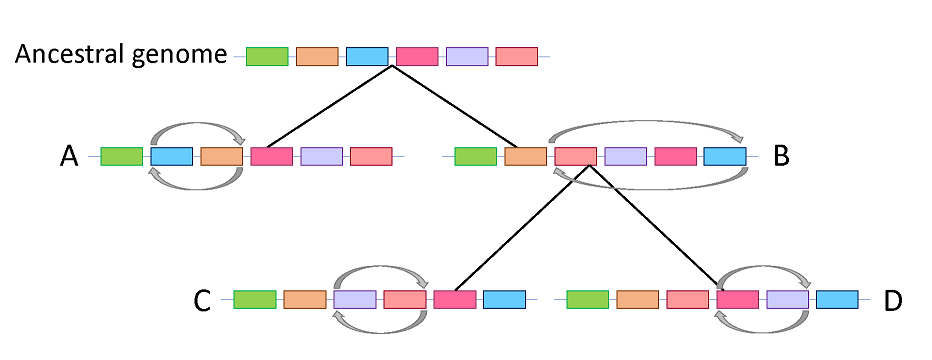
Genome rearrangements are the changes that occur in the genome of every organism which helps them to adapt to the environment. There are different types of genome rearrangements but in bacteria the most common is inversion (part of genome is reversed). Helicobacter pylori is a bacterium which resides in the human stomach and has a characteristic of changing its genomic structure in order to survive in the acidic environment. It is important to identify the genome rearrangements as they can provide information regarding the evolutionary history of the organisms (Figure). Evolutionary history of the organisms can be tracked much farther back using genome rearrangements as they are rare compared to the other genetic changes occurring in the genome. Several approaches have been proposed to identify the genome rearrangements but most uses pairwise comparison and consider only the fully conserved genes. However, multiple genome comparison is required to identify the changes and provide the information about the evolutionary events.
In order to identify the genome rearrangements while comparing multiple genomes of closely related strains, we have developed a method which is different from the previous ones as it takes into account the missing genes also. Almost conserved gene clusters were used to generate the gene order of each genome. Inversions were identified by comparing the gene orders of multiple genomes. Several inversions were identified, few were found to be related to geographical region-related. Some inversions were found to be associated with disease states. As H. pylori has evolved with the global human migration, identifying the inversions may provide insight into the patterns of migration.
I had visited National Cancer Institute in USA from September 6th to October 24th under SOKENDAI Student Dispatch Program 2019. I am working on Helicobacter pylori Genome Project ( Hp GP) in collaboration with the group in USA, during my stay I worked on assigning the protein names (called annotations) for the Helicobacter pylori strains. For this I have created the library which will provide the consistent annotations for the research community.
Genetics, Mehwish Noureen
