March 05, 2020
Application of cell-free DNA in monitoring graft injury and rejection in kidney transplant patients.
SOKENDAI Student Dispatch Program program year: 2019
Nguyen Thanh Phuong, Genetics
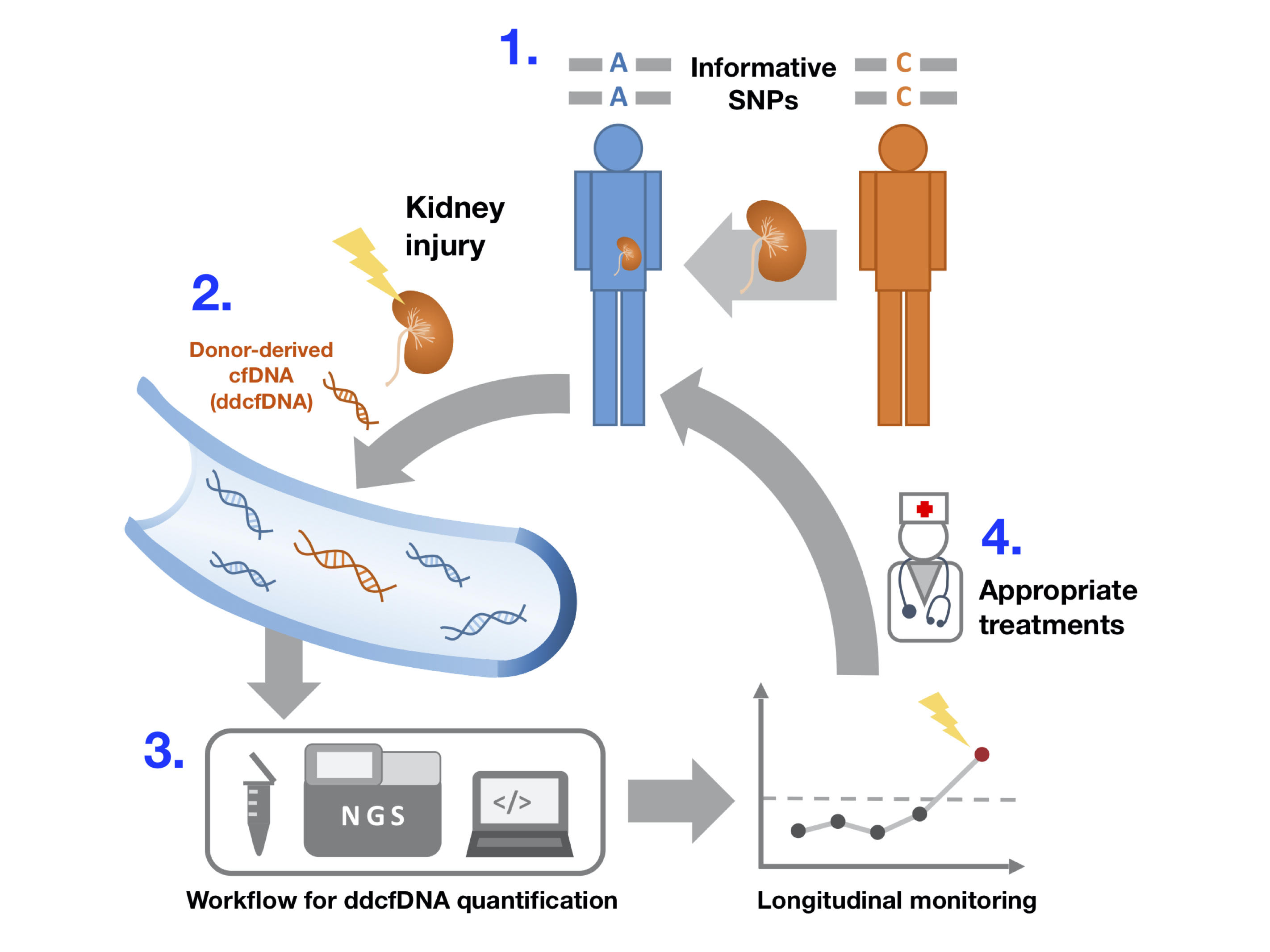
1. Determining genotypes of Donor and Recipient; 2. Injury or rejection of transplant kidney releases donor DNA (ddcfDNA) into blood stream; 3. Cell-free DNA extraction, Next-generation sequencing and computational methodology help quantify ddcfDNA level; 4. Regular monitoring of this level help doctor understand the well-being of transplant kidney and take responsive action if necessary.
Monitoring graft conditions in transplant recipient requires intensive follow-up medical examinations. In case of kidney transplantation, organ integrity is conventionally evaluated by biopsy combined with measurement of serum markers. Although biopsy gives accurate visual evidences for diagnosis, it is an invasive technique and hence not ideal to be performed multiple times on patient. Other serum markers are mostly based on the functionality of kidney and generally can not tell us about rejection damages at cellular level. Our ultimate target is to develop diagnostic method using a new biomarker based on genetic information which only employs blood collected from kidney transplant recipients. We sought to resolve 3 main tasks in this project: Developing and optimizing our methodology, confirming the potential of this biomarker based on clinical data, and comparing it with other conventional serum markers.
Basically, except for the case of monozygotic twin siblings, when an organ is received from a donor, the immune system of recipient always recognizes it as "foreign" and evoke immune responses by various mechanisms to "attack" that organ. As a consequent, injury of transplanted organ releases a portion of its "foreign" DNA into blood circulation in the form of cell-free DNA (termed donor-derived cell-free DNA or ddcfDNA for short). But, the question is how to recognize this cell-free DNA from donor? Since genomes of different individuals in population differ in the type of nucleotide at specific positions, called single nucleotide polymorphism sites (SNP sites), we exploited these SNP sites to know which DNA comes from donor and also quantify its level compared to the total cell-free DNA in recipient. The level of this donor-derived DNA can be directly translated into the degree of damage of transplanted organ.
We collected more than 300 plasma samples from 39 kidney transplant patients with extensive follow-up timepoints. We successfully established a rapid and efficient sequencing methodology on extracted cell-free DNA which specifically targets 1000 selected SNP sites. To confirm the clinical value, we observed that ddcfDNA levels showed indicative signals for all rejection episodes and other various types of kidney injury, which was not reported by other conventional markers. Additionally, we found evidences showing that ddcfDNA level was more sensitive in reflecting immunosuppressants dosage administered on recipients. In conclusion, our study demonstrated ddcfDNA level as an advantage non-invasive marker for monitoring various aspects of graft health, its integration with conventional clinical markers can provide better diagnosis and support decision making for doctors in kidney transplant healthcare. We hope to extend this study in other organ transplantation and adopt large-scale clinical application in the near future.
Achievements through the SOKENDAI Dispatch program
I am truly grateful to receive the financial support from SOKENDAI Dispatch program to introduce above-mentioned study in American Society of Human Genetics Annual Meeting (ASHG 2019) which was held from 15th to 19th October, 2019 in Houston, USA. It was an honor for me to be selected as platform speaker on Session 97: "Chromosome to Cell-free DNA: Balancing Genetic Contributions". During my visit to ASHG 2019, I experienced the dynamics and fascinating spirit of this research community. I learned much about cutting edge technologies and current research trends from leading institutes of USA in the field of human genetics. With oral presentation and discussion, I had chance to expose my research and receive many valuable comments and suggestion.
Lastly, I would like to express my sincere gratitude toward SOKENDAI Dispatch program for providing me this grant. I would like to acknowledge my supervisor, all laboratory members, collaborating researchers and doctors, and especially to all patients who participated in our study.
Genetics, Nguyen Thanh Phuong
